Pitfalls of ascertainment biases in genome annotations—computing comparable protein domain distributions in eukarya
DOI:
https://doi.org/10.11113/mjfas.v10n2.57Keywords:
protein domains, HMM models, GO classification, functional genomeAbstract
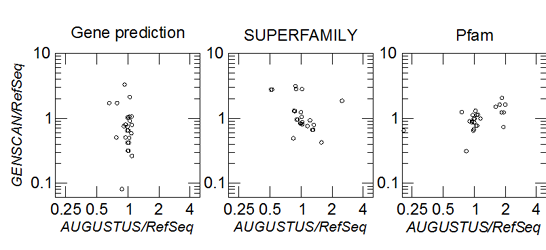
Most investigations into the large-scale patterns of protein evolution are based on gene annotations that have been compiled in reference databases. The use of these resources for quantitative comparisons, however, is complicated by sometimes vast differences in coverage. More importantly, however, we also observe substantial ascertainment biases that cannot be removed by simple normalization procedures. A striking example is provided by the correlations between protein domains. We observe that statistics derived from different computational gene annotation procedure show dramatic discrepancies, and even qualitative changes from negative to positive correlation, when compared to statistics obtained from annotation databases.
________________________________________
GRAPHICAL ABSTRACT
References
K. Forslund, E.L.L. Sonnhammer, Bioinformatics 24 (2008) 1681.
G. Apic, J. Gough, S.A. Teichmann, J. Mol. Biol. 310 (2001) 311.
C.A. Orengo, J.M. Thornton, Annu. Rev.Biochem. 74 (2005) 867.
M. Buljan, A. Bateman, Biochem. Soc. Trans. 37 (2009) 751.
A.D. Moore, A.K. Björklund, D. Ekman, E.Bornberg-Bauer, A. Elofsson, Trends. Biochem. Sci. 33 (2008) 444.
E. Koonin, L. Aravind, A. Kondrashov, Cell 101 (2000) 573.
E. Bornberg-Bauer, A.K. Huylmans, T. Sikosek, Curr. Opin. Struct. Biol. 20 (2010) 390.
C.M. Zmasek, A. Godzik, Genome Biol. 12 (2011) R4.
K. Mahmood, G. Webb, J. Song, J. Whisstock, A. Konagurthu, Nucleic Acids Res. 40 (2012) e44
K.M. Kim, G. Caetano-Anolles, BMC Evol. Biol. 11 (2011) 140.
S.J. Prohaska, P.F. Stadler, D.C. Krakauer, J. Theor. Biol. 265 (2010) 27.
S. Yang, P.E. Bourne, PLoS ONE 4 (2009) e8378.
W. Stefan, A. Eivind, BMC Evol. Biol., 5 (2005) 24.
A.D. Moore, S. Grath, A. Schüler, A.K. Huylmans, E. Bornberg-Bauer, Biochim.Biophys.Acta1834 (2013) 898.
K. Nowick, A.T. Hamilton, H. Zhang, L. Stubbs, Mol. Biol. Evol. 27 (2010) 2606.
D.A.de Lima Morais, H. Fang, O.J. Rackham, D. Wilson, R. Pethica, C. Chothia, J. Gough, Nucleic Acids Res. 39 (2011) D427–D434.
C. Burge, S. J. Karlin, Mol. Biol. 268 (1997) 78.
C.B. Burge, S. Karlin, Curr. Opin. Struct. Biol.8 (1998) 346.
I. Korf, BMC Bioinformatics, 5 (2004) 59.
W. Miller, K.D. Makova, A. Nekrutenko, R.C. Hardison, Ann. Rev. Genomics Hum. Genet. 5 (2004) 15.
T. Hubbard, D. Barker, E. Birney, G. Cameron, Y. Chen, L. Clark, T. Cox, J. Cuff, V. Curwen, T. Down, R. Durbin, E. Eyras, J. Gilbert, M. Hammond, L. Huminiecki, A. Kasprzyk, H. Lehvaslaiho, P. Lijnzaad, C. Melsopp, E. Mongin, R. Pettett, M. Pocock, S. Potter, A. Rust, E. Schmidt, S. Searle, G. Slater, J. Smith, W. Spooner, A. Stabenau, J. Stalker, E. Stupka, A. Ureta-Vidal, I. Vastrik, M. Clamp, Nucleic AcidsRes. 30 (2002) 38.
H.Korenaga, T.Kono, M.Sakai, Cytokine 59 (2012) 541.
A.A.Parikesit, P.F.Stadler, S.J.Prohaska, Genes 2 (2011) 912.
M.Stanke,S.Waack, Bioinformatics 19 (2003) ii215–ii225.
M.Stanke,O.Schöffmann,B.Morgenstern,S.Waack,BMC Bioinformatics 7 (2006) 62.
M.Stanke, M.Diekhans, R.Baertsch, D.Haussler,Bioinformatics, 24 (2008) 637.
M.Stanke, Augustus Training Manual: http://bioinf.uni-greifswald.-de/augustus/binaries/tutorial/training.html. ioinformatics Group, University of Greifswald. (2011).
A.Quinlan, I. Hall, Bioinformatics 26 (2010) 841.
S.L.Baldauf, J. Syst. Evol.46 (2008) 263.
B.Dilts, United States Patent Application Publicatin No: US 2012/0081389 (LucidChart) (2012), Technical report toLucidChart LLC.
S.R. Eddy, PLoS Comp. Biol. 7 (2011) e1002195.
The Gene Ontology Consortium. Nat Genet. 25 (2000) 25.
M.Punta, P.C.Coggill, R.Y.Eberhardt, J.Mistry, J.Tate; C.Boursnell, N.Pang, K.Forslund, G.Ceric, J.Clements, A.Heger, L.Holm, E.L.Sonnhammer, S.R.Eddy, A.Bateman,R.D.Finn, Nucleic Acids Res. 40 (2012) D290–D301.
Y.Niimura, M.Nei,PLoS ONE 2 (2007) e708.
K.D.Pruitt, T.Tatusova, D.R.Maglott, Nucleic Acids Res. 33 (2005) D501–D504.
A.A.Parikesit, P.F.Stadler, S.J.Prohaska, German Conference on Bioinformatics 2010,D.Schomburg, A.Grote,Eds. Gesellschaft fürInformatik: Bonn, 2010. Vol. P-173, Lecture Notes in Informatics, pp.93–102.
G.C. Conant, G.P.Wagner, P.F.Stadler, Mol. Phylog. Evol. 42 (2007) 298.
W.C. Wong, S.Maurer-Stroh, F.Eisenhaber, PLoSComput. Biol. 6 (2010) e1000867.
S.Michaeli, Future Microbiol. 6 (2011) 459.
S.Thomas, A.Green, N.R.Sturm, D.A.Campbell, P.J.Myler,BMC Genomics 10 (2009) 152.
F. Lu, H.Jiang, J.Ding, J.Mu, J.G.Valenzuela, J.M.C.Ribeiro, X.z.Su,BMCGenomics 8 (2007) 255.
L.M.Iyer, V.Anantharaman, M.Y.Wolf, L.Aravind, Int. J.Parasitol. 38 (2008) 1.
J.Eisen, et al. PLoS Biol. 4 (2006) e286.
R.D. Adam,A.Nigam, V.Seshadri, C.A.Martens, G.A.Farneth, H.G.Morrison, T.E.Nash, S.F.Porcella, R.Patel,BMC Genomics 11 (2010) 424.
I.A.Drinnenberg, D.E.Weinberg, K.T.Xie, J.P.Mower, K.H.Wolfe, G.R.Fink, D.P.Bartel, Science 326 (2009) 544.
R.Melzer, G.Theissen, MethodsMol. Biol. 754 (2011) 3.
E.Shelest, FEMSMicrobiol.Lett. 286 (2008) 145.
B.Banfai, H.Jia, J.Khatun, E.Wood,B.Risk, W.E.GundlingJr., A.Kundaje,H.P.Gunawardena, Y.Yu, L.Xie, K.Krajewski, B.D.Strahl, X.Chen, P.Bickel, M.C.Giddings, J.B.Brown, L.Lipovich, Genome Res. 22 (2012) 1646.
A.R.Kersting, E.Bornberg-Bauer,A.D.Moore, S.Grath, Genome Biol.Evol. 4 (2012) 316.
A.D. Moore, E.Bornberg-Bauer, Mol. Biol.Evol. 29 (2012) 787.
S.Yang, R.F.Doolittle, P.E.Bourne, Proc.Natl. Acad. Sci. USA 102 (2005) 373.
O. Niehuis, G.H. Hartig, S.Garth, H.Pohl,J.Lehmann, H.Tafer, A. Donath, V.Krauss, C.Eisenhardt, J.Hertel, M.Petersen, C.Mayer, K.Meusemann, R.S.Peters, P.F.Stadler,R.G.Beutel, E.Bornberg-Bauer, D.D.McKenna, B.Misof,Current Biol. 22 (2012)1309.
A.A. Parikesit, Supplemental Data: http://www.bioinf.uni-leipzig.-de/supplements/12-007. (2012) Bioinformatics groups, University of Leipzig.



















